Dr Amin Turki was proud to present results from research conducted
within the HARMONY Platform at the European Hematology Association (EHA) Annual Congress, celebrated in Milan.

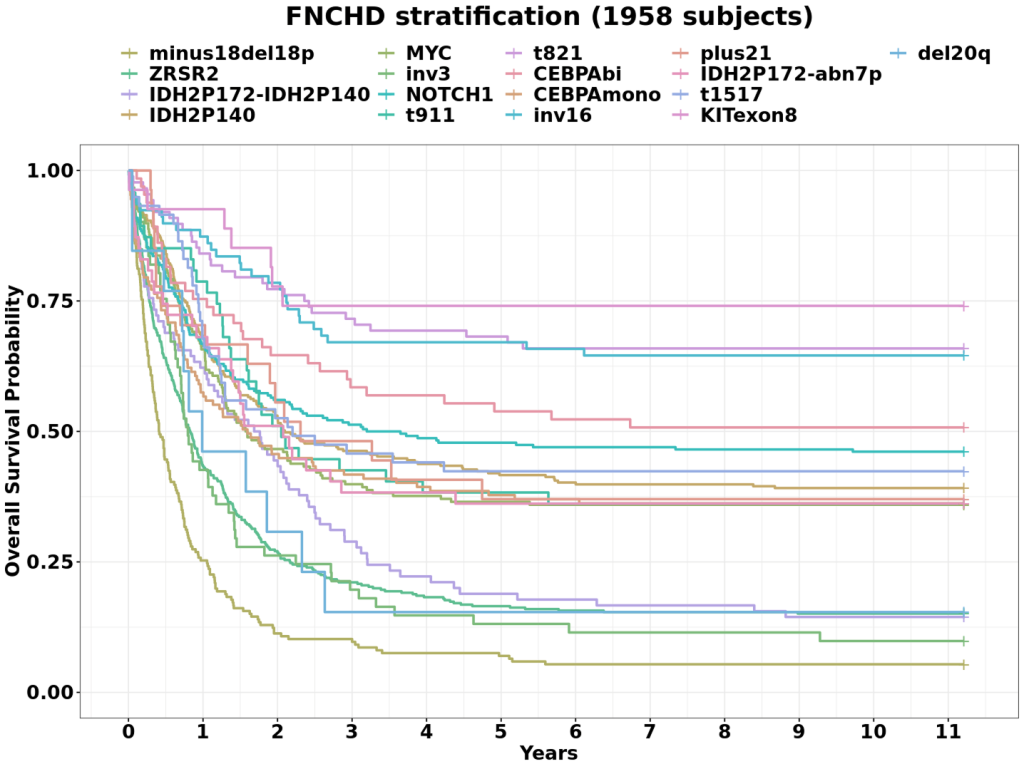
Fig 1 of the presentation.
Every year, the European Hematology Association (EHA) holds its Annual Congress, bringing together clinicians, researchers, and other stakeholders in hematology. This year, the 30th edition, took place in Milan, Italy, between the 12th and the 15th of June 2025.
Under the slogan “Connect. Enrich. Progress”, thousands of experts participated in symposia, meetings and other scientific presentations.
Amin Turki, MD, PhD, of University Hospital Bochum in Germany, participated in the EHA Plenary Abstracts session “Genetic aberrations and AML risk scoring” to present the work developed within the HARMONY Platform: “ Development and Independent Validation of Unsupervised Genomic Classification of Patients With Acute Myeloid Leukemia Using Hierarchical Dirichlet Mixture Models: A HARMONY Alliance Study”.
During the session, he presented new insights into AML classification through machine learning: using Dirichlet mixture models, his team identified 17 distinct biological clusters, each linked to differences in overall survival extending beyond 11 years of follow-up, which, in his words, “has the potential to further personalize outcome prediction”. He also emphasized the HARMONY Alliance Foundation’s openness to collaboration, highlighting the opportunity to unlock deeper genetic insights by analyzing even larger datasets.
Relevant links:
- This presentation will be accessible for on-demand viewing from June 18 to August 15, 2025 on the EHA Congress platform.
- The abstract can be accessed on the following link: https://library.ehaweb.org/eha/2025/eha2025-congress/4159225/amin.turki.development.and.independent.validation.of.unsupervised.genomic.html
Summary of the presentation:
Acute myeloid leukemia (AML) is the most common acute leukemia, but it is heterogenous in biology and outcomes, with disease-related factors encoded in molecular- and cytogenetics. To tackle this heterogeneity, we used an unprecedently large dataset of AML patientes developed by HARMONY project. On this basis, we redesigned the post-processing of the Dirichlet method to achieve a fully unsupervised genomic classification.
As a result, we identified 17 gene clusters composed of several driver mutations with distinct co-mutational patterns to characterize the AML landscape. In patients treated with intensive chemotherapy (n=4836), these 17 genetically defined components associated with significantly distinct overall survival (OS, p<0.0001).
Both the distribution of components as well as the significant differences in OS (p<0.001, Fig. 1) and relapse free survival were independently validated. Other previously-described components were confirmed. We further detected a novel component that prioritized deletions –in 20q, which are more common in myelodysplastic syndromes. This is the largest study of its kind and provides important insights into the heterogeneity of AML genetics adding new classes and subdivisions of existing classes, while maintaining the concept of driver and passenger mutations. Its independent multicenter validation underlines both generalizability and robustness of this new method.
